Building CalculiX with the PaStiX solver without CUDA
The CalculiX solver package on FreeBSD is compiled with the SPOOLES (SParse Object Oriented Linear Equations Solver) library by default. This is also the case in many Linux distributions because SPOOLES is free, while faster solvers like PARADISO are proprietary.
SPOOLES is relatively fast compared to the built-in iterative solver, its most fundamental limitation is that the data must fit in RAM.
However a patched version of the PaStiX solver has been integrated with CalculiX. It is often faster than SPOOLES and can use a GPU using the CUDA library.
So I wanted to try CalculiX built with PaStiX. There is just one slight issue; CUDA is not supported on FreeBSD. And I use Intel or AMD graphics because those are supported by open-source drivers.
Therefore I needed to build PaStiX without CUDA support. This turned out to be slightly more complicated then expected.
Currently, CalculiX used a modified version of PaStiX that you can find at https://github.com/Dhondtguido/PaStiX4CalculiX. This is based on an older version of PaStiX. And even though it has a configuration option to build without CUDA, that doesn’t work. Basically, even if you switch CUDA off, there is still some CUDA-specific stuff left. I was halfway through patching all that, when I found that github user Kabbone had already done all of that: https://github.com/Kabbone/PaStiX4CalculiX/tree/cudaless. So that is the version I’m using.
Support for CalculiX in mainline PaStiX is being worked on, but will certainly not arrive before PaStiX 6.5. Meanwhile Kabbone has a branch in a fork of CalculiX that builds with PaStiX 6.4. I haven’t tried that yet.
Prerequisites
These build instructions were written for FreeBSD. With some changes they should be usable on other UNIX-like systems like Linux and MacOS, and maybe even MSYS2.
Building software on ms-windows is such an exercise in self-flaggelation to me that I gladly avoid it. The contents of the patches and scripts might still be useful in this case. The latter as a general guideline.
The following software packages or ports are required for a successful build:
- GNU make (called
gmakeon FreeBSD, usuallymakeon Linux) - A Fortran compiler. Here GNU fortran in the form of
gfortran13is used. (On Linux it’s probably just calledgfortran) - A C compiler. Here
gcc13is used. (orgccon Linux) - GNU autotools/automake/libtool
- cmake
- bison
- pkg-config
- Python 2.7 (for PaStiX code expansion)
- Python 3
For the build I set up the following directory tree:
. ├── bin ├── distfiles ├── examples ├── include ├── lib ├── libexec ├── logfiles ├── patches ├── share ├── source └── unused
All the distribution files of known working versions are stored in distfiles.
The needed patches are stored under patches.
Log files of the builds are saved under logfiles.
The builds are done under the source directory.
Scripts and patches that are not in use anymore are stored under unused.
The other directories (examples, include, lib, libexec,
share) are locations for the libraries to be installed in.
The root directory contains the scripts used for the build. This whole set of distfiles, patches and shell scripts can be found on github.
License
The files in distfiles are under their respective licences.
The materials that I wrote are hereby placed in the public domain.
Build
A whole stack of libraries needs to be built in order to be able to build CalculiX with PaStiX. Building CalculiX itself is the last step. In order:
- SPOOLES 2.2
- OpenBlas 0.3.28
- arpack-ng 3.9.1
- hwloc 2.11.2
- mfaverge-parsec-b580d208094e
- scotch 7.0.6
- PaStiX4CalculiX (cudaless branch from https://github.com/Kabbone/PaStiX4CalculiX)
- CalculiX 2.22
Before the build proper is started, the clean.sh script is started from
the root directory of the repository.
This will ensure a clean build.
Configuration
Except for parsec, all other libraries are available in the FreeBSD ports
tree.
However, their standard configurations are generally different than those required for
this build.
Therefore all the libraries are only built as static libraries so the code
is linked into CalculiX so we don’t have multiple configurations of shared
libraries around; that way lies madness.
This can be done with environment variables like NO_SHARED=1, or it can be
as a configuration options --disable-shared and --enable-static or
-DBUILD_SHARED_LIBS=OFF.
In several of the build scripts the variable PREFIX is defined as the
output of the pwd command.
This is used as the location where the built libraries are to be installed,
and where other libraries can be found.
So it is important that all the build scripts are called from the directory in
which they are located.
The compilers to use are often specified as environment variables:
CC=gcc13for the C compiler,CXX=g++13for the C++ compiler,FC=gfortran13for the fortran compiler,AR=gcc-ar13for the archiver.
For both hwloc and PaRSEC I explicitly added libexecinfo and libpciaccess to
the configuration.
SPOOLES
It is good to have SPOOLES available next to PaStiX as a back-up. Also, SPOOLES is generally faster for small calculations or eigenfreqency calculations.
For building SPOOLES, I basically applied the same patches as in the FreeBSD
ports tree.
Patches that I added were to ETree/src/transform.c and
Utilities/src/iohb.c to fix compiler warnings.
And of course Make.inc was patched to select the compiler and build options.
The script 01_build_spooles.sh is used to build SPOOLES. It is called as follows, to also redirect the output to a log file:
sh 01_build_spooles.sh |& tee logfiles/spooles.log
OpenBLAS
The routines in OpenBLAS, which not only includes BLAS but also LAPACK, are the core routines used in the solver.
The script 02_build_openblas.sh is used to build OpenBLAS:
sh 02_build_openblas.sh |& tee logfiles/openblas.log
This library does not require patches.
It just needs to be configured for building and installing.
The env program is used to communicate the configurations as environment
variables to gmake.
The most important configurations are:
NO_SHARED=1for a static library.INTERFACE64=1for 8-byte integers.USE_THREAD=0because CalculiX wants a single-threaded library.USE_LOCKING=1since CalculiX itself is threaded. Leaving this setting out results in a non-working CalculiX!.
Below is an example of what an example result looks like when OpenBLAS is configured without locking:
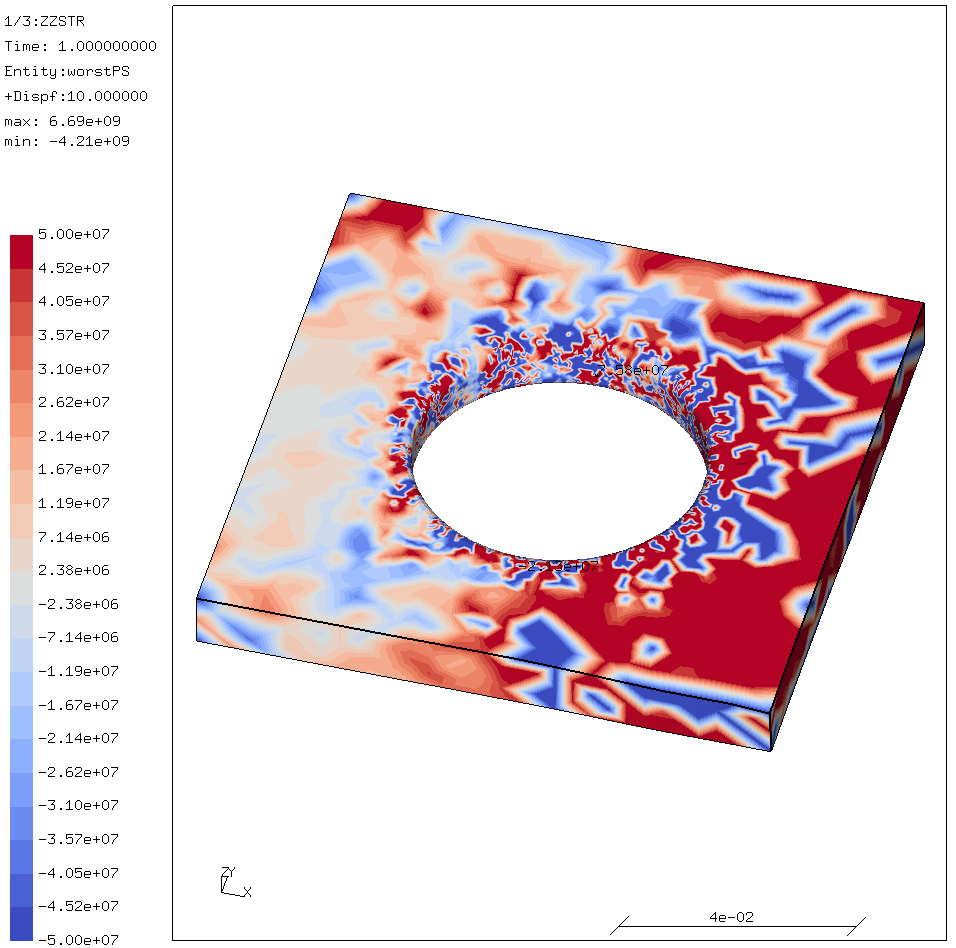
This is what it is supposed to look like:
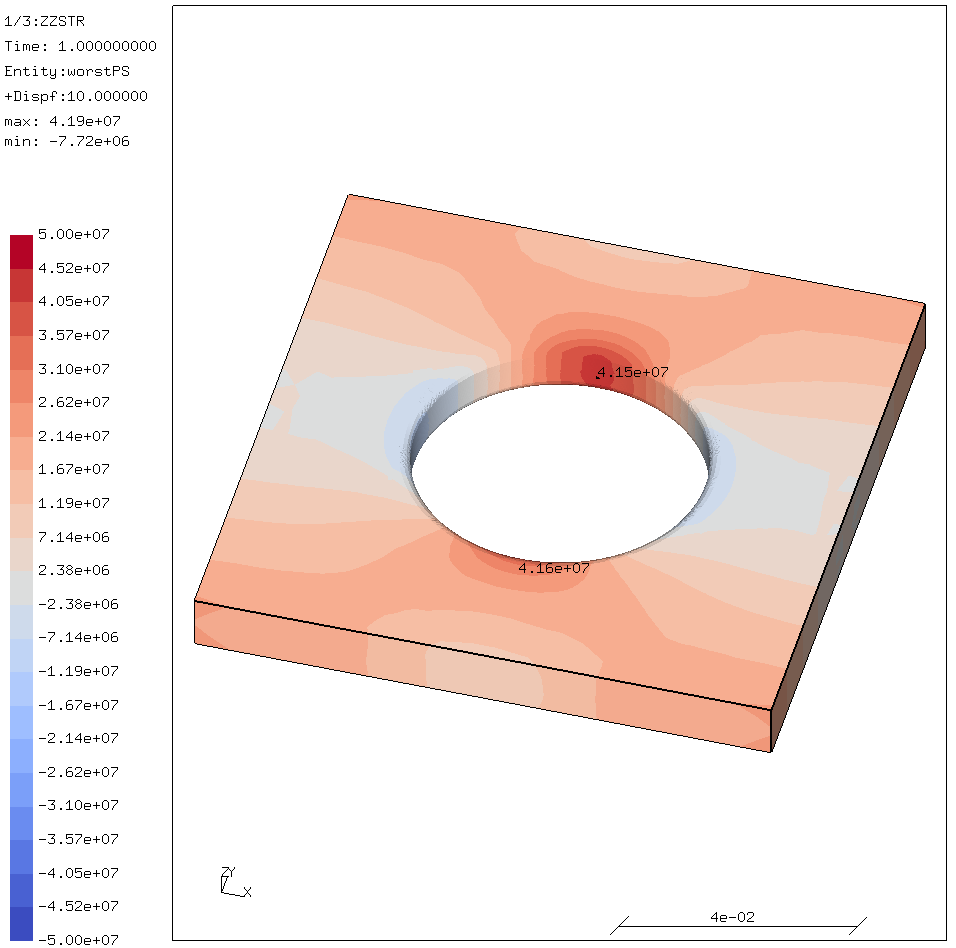
There are two other settings that one might change;
BUFFERSIZE=25increases the internal buffer from 32 MiB to 1 GiB.DYNAMIC_ARCH=1will build OpenBLAS for all revelant processor types and chooses the right one at runtime. Disabling this will make the build a lot faster but also restricts CalculiX to running on the same processor generation as it is built on (or a later one).
ARPACK
The ARPACK library is used to solve eigenvalue problems. The original library is no longer maintained, so we will use the fork arpack-ng.
The build script is 03_build_arpack.sh:
sh _03_build_arpack.sh|& tee logfiles/arpack.log
It is configured with the environment variable INTERFACE64=1 to use 8-byte
integers.
The compilers to use are configured like this as well.
Important options given to the configure script are:
--with-blas=-lopenblasand--with-lapack=-lopenblasto tell it to use OpenBLAS.--enable-staticand--disable-sharedfor obvious reasons.
hwloc
The hwloc library is used as a portable abstraction for the topology of modern hardware.
Most of the possible extras were disabled in the build script 04_build_hwloc.sh, since we are only interested in the library:
sh 04_build_hwloc.sh|& tee logfiles/hwloc.log
PaRSEC
PaRSEC is used as a scheduling framework for multitasking.
The build script is 05_build_parsec.sh:
sh 05_build_parsec.sh|& tee logfiles/parsec.log
In this build I had to fix two header file names. This was done using sed.
And I added a small patch to the parsec_bindthread that I found in one of
the forum threads, IIRC.
Since CUDA is not used, it is possible to skip this library, and let CalculiX statically configure tasks. From what I’ve read, this might be suboptimal and/or require patching of CalculiX. For that reason I just included it.
scotch
Either scotch or METIS can be used by PaStiX for ordering of sparse matrices.
However, I could not get PaStiX to detect METIS properly.
And scotch is supposed to be faster, so scotch it is.
If your CPU has more or less than 4 cores, you might want to change
-DSCOTCH_PTHREAD_NUMBER=4 in patches/scotch/Makefile.inc.RFS accordingly
before calling the build script 06_build_scotch.sh
sh 06_build_scotch.sh | & tee logfiles/scotch.log
PaStiX4CalculiX
As mentioned before, this is a custom version of PaStiX modified to work with CalculiX and without CUDA.
This library contains one version of the code (for complex numbers) which is
then converted into other versions (float and double for example) at
compile time.
This older version of PaStiX requires Python 2.7 to do this.
Python 3 will not work!
Note that you will have to adapt the build script 07_build_pastix_kabbone.sh
if Python 2.7 is not in /usr/local/bin/python2.7:
sh 07_build_pastix_kabbone.sh|& tee logfiles/pastix.log
CalculiX
Finally it it time to build CalculiX using 08_build_calculix.sh:
sh 08_build_calculix.sh|& tee logfiles/calculix.log
A couple of patches are used to silence warnings. The Makefile is customized to configure the build and link to the correct libraries.
The build script installs the stripped binary in ${PREFIX}/bin as
ccx_i8, to distinguish it from the regular FreeBSD package which uses
ccx.
After testing I tend to copy it to /usr/local/bin/ccx, overwriting the
executable from the package.
History
| Date | Change |
| 2024-04-25 | Initial version. |
| 2024-12-30 | Update CalculiX 2.22, Openblas 0.3.28, hwloc 2.11.2, scotch 7.0.6 |
For comments, please send me an e-mail.
Related articles
- Element names in Calculix
- FEA based on STEP geometry using gmsh and CalculiX
- Reading CalculiX mesh with Python
- Folded leaf spring ball joint flexure
- Creating a rectangular tube in CalculiX